About
Authors
Amir Reza Ghods
Yuguang Zhang
Information
Type: Infrastructure Service
License: AGPLv3
Timeline: 2013-present
Institution: University of Waterloo
Links
Main Site (datamill.uwaterloo.ca)
Project Site (uwaterloo.ca)
Source Code (bitbucket.org)
Motivation
From their project page:
Empirical systems research is facing a dilemma. Minor aspects of an experimental setup can have a significant impact on its associated performance measurements and potentially invalidate conclusions drawn from them. Examples of such influences, often called hidden factors, include binary link order, process environment size, compiler generated randomized symbol names, or group scheduler assignments. The growth in complexity and size of modern systems will further aggravate this dilemma, especially with the given time pressure of producing results. So how can one trust any reported empirical analysis of a new idea or concept in computer science?
DataMill facilitates producing robust, reliable, and reproducible results. The infrastructure incorporates the latest results on hidden factors and automates the variation of these factors.
Rubric
✔ - Yes
✗ - No
○ - Yes, but with concession
· - Inapplicable
? - Unknown
| Infrastructure | ||
| Self-Hosting | ✗ | One cannot self-host a complete DataMill cluster. There are some instances where a worker node will communicate directly with a master node. These are hard-coded and there is no support or documentation given on how to establish either a master node or a worker node for that matter. |
| Provides Metadata | ✔ | |
| Provides Hardware Diversity | ✔ | |
| Dispatches Work to Cloud Machines | ✗ | |
| Provides a Web Portal | ✔ | |
| Provides Performance Monitoring | ❓ |
| Capabilities | ||
| Runs Code | ✔ | General Purpose: Can run anything that can be wrapped in several bash scripts and works on the infrastructure |
| File Storage | ✔ | |
| Collaboration Controls | ❓ | |
| Provides Citations | ✗ | |
| Interactive Graphing | ✗ | You can download the resulting files. The service provides to further mechanism to read or interpret the result. |
| Can Combine Objects | ✗ | One package is executed. There is no method of combining existing artifacts and running them together or chaining them together. |
| Can Archive/Run GUI Tools | ✗ | Only non-interactive command line scripts. |
| Can Hook to External Services | ✗ |
| Access | ||
| Public view of object | ✗ | All views of an object require logging in. Accounts are only manually given out. |
| Access Permissions for Editing | ❓ | |
| Access Permissions for Reading | ❓ | |
| Access Permissions for Anon Review | ❓ |
| Provenance | ||
| Search | ✔ | |
| Globally Unique Identifiers for Projects | ✗ | Only locally unique identifiers. |
| Provides URL to Project / Data | ✔ |
| Governance | ||
| Open Source | ✔ | AGPLv3 |
| Allows Modification / Redistribution | ✔ | |
| Has a Free-to-Use Package | ✗ | |
| Has a Student Package | ✔ | Only students and manually verified university researchers may make use of the service. |
| Has a Paid Package | · |
Walkthrough
DataMill is a service that allows you to attach machines to use as worker nodes and create experiments which are to be scheduled and executed among those workers. To do this, assuming there are already workers available and established, is to package up your work in a particular way and upload it to the DataMill service.
To do that, you would write three scripts: setup.sh, run.sh, and collect.sh. Some examples (mostly the same as ones given
in their paper) follow:
setup.sh
This script will be executed when the job is being prepared to run on the worker node.
#!/bin/sh
cd /dry
chmod +x dry.c # Apparently this is a compiled file?? I think you would build here.setup.sh
This script will execute the job. Any measurements that can be used on the worker node will measure whatever this script runs.
#!/bin/sh
cd /dry
./dry.c >> results 2>&1collect.sh
This cleans up and writes out any information the experimenter wants recorded. The standard output when this runs will be recorded separately and made available to download. Generally, you want this to be the data you want to pull out of the experimentation.
#!/bin/sh
results=dhrystone-`date +%Y%m%d_%H%M`.tar.gz
tar czf $results /dry/results > /dev/null 2>&1
echo $resultsUploading
Once you have these three scripts (which you can test on a test virtual machine image) you can create an experiment on the service. Assuming you have an account (you must be affiliated with a university and your account will be manually moderated), you can navigate to "Experiments" to find a "Create new experiment" button.
From here, you can specify the name and description of the experiment you will upload. You can specify a replicate count which suggests how many times the experiment will run. Tags can be recorded to help group and search for related experiments later.
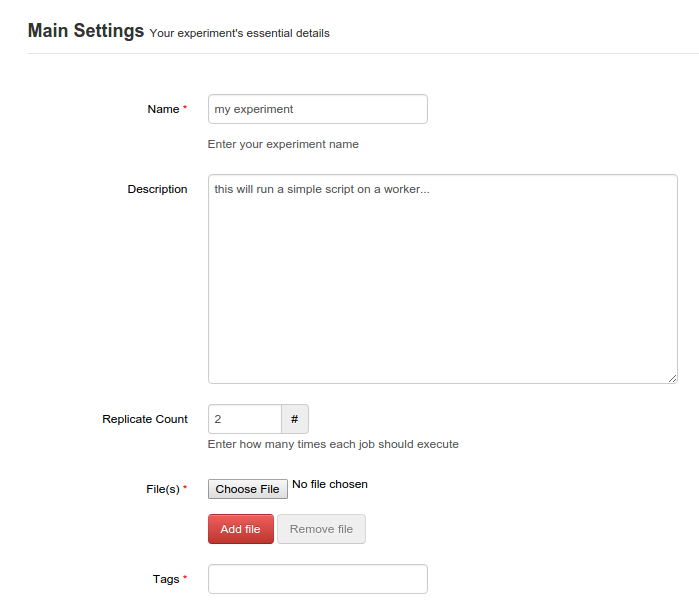
Here you upload your scripts and data as a tarball. Simple metadata for the running experiment can be provided. You can specify you want to replicate the experiment a number of times.
The next section allows the experimenter to specify the hardware requirements for the experimentation. By default, the entire ranges are specified. You can slide the bar from either end to restrict the possible hardware choices you will have. At the bottom, the workers that can accommodate the requirements are highlighted.
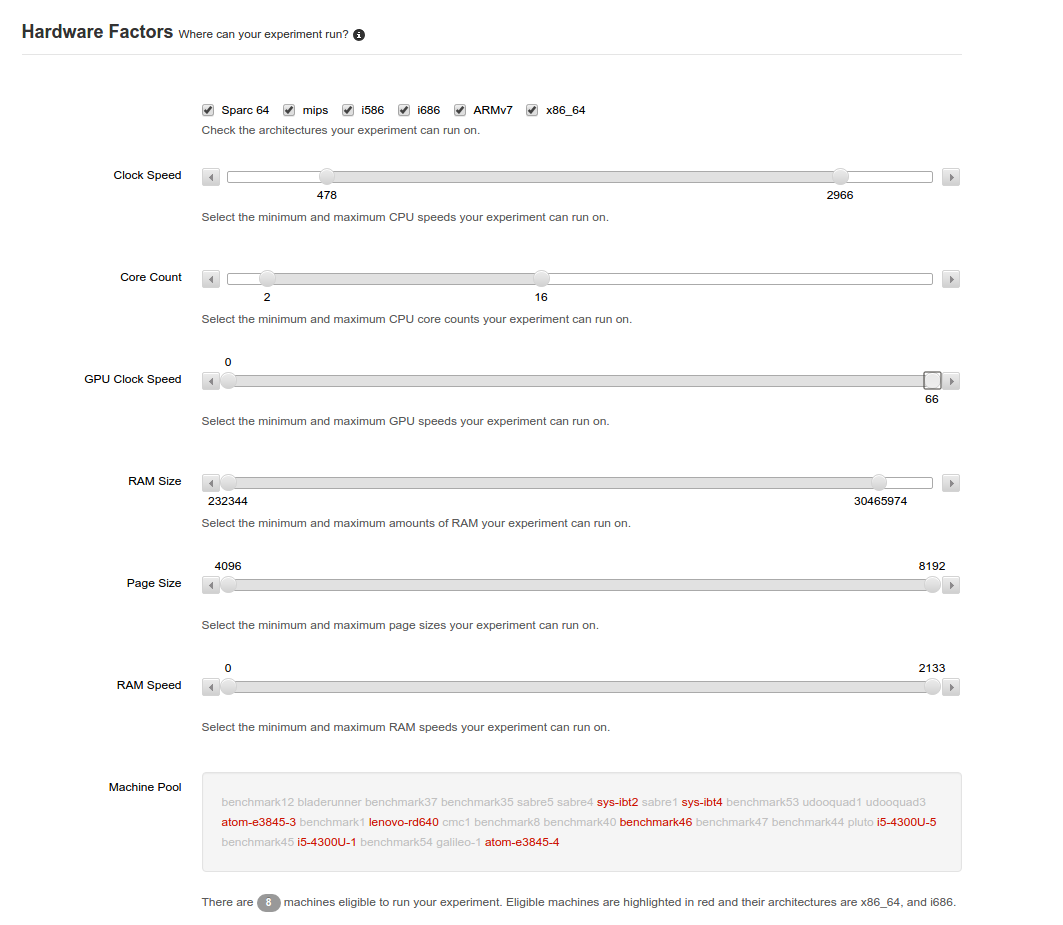
Ranged sliders allow one to determine which known worker machines can be used to run the experiment.
Once the requirements have been determined and that availability of workers assured, the next section
is used to toggle different features that can be used with the experiment.
These are well-known factors where the system can automatically influence the experiment.
For instance, different optimization flags can be toggled which will replicate and run the experiment
for each option. If you toggle ‑O2 and ‑O0 and also have both gcc and llvm, then the experiment
will run 4 times with each permutation of options. Each time, it will influence the standard environment
variables CC etc to reflect these choices. It is up to the scripts to react to those variables if it
is not automatic (for instance, within a Makefile).
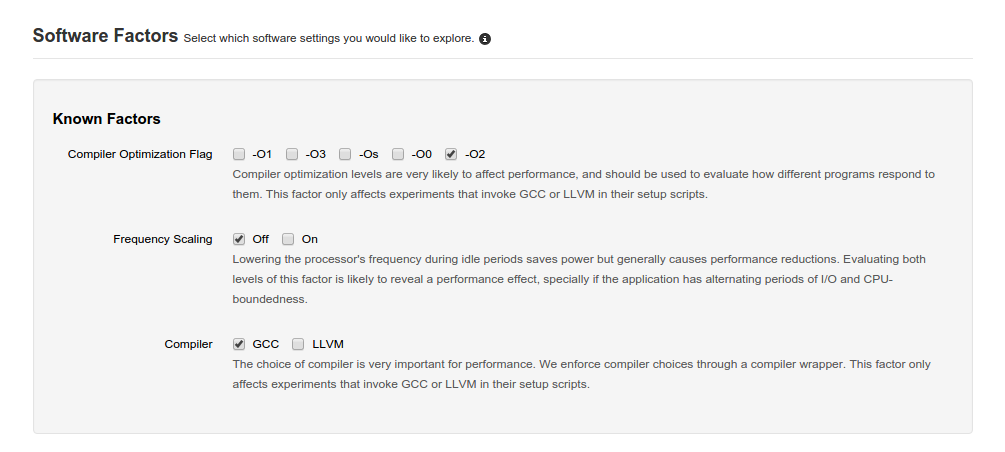
Other factors can be toggled here that are specific to well-known applications available in the infrastructure such as compile flags.
There are also hardware factors where an experimenter can select different hardware configurations and replicate the experiment across different workers and architectures. This way, one can control for the hardware or do a type of continuous testing across different platforms.
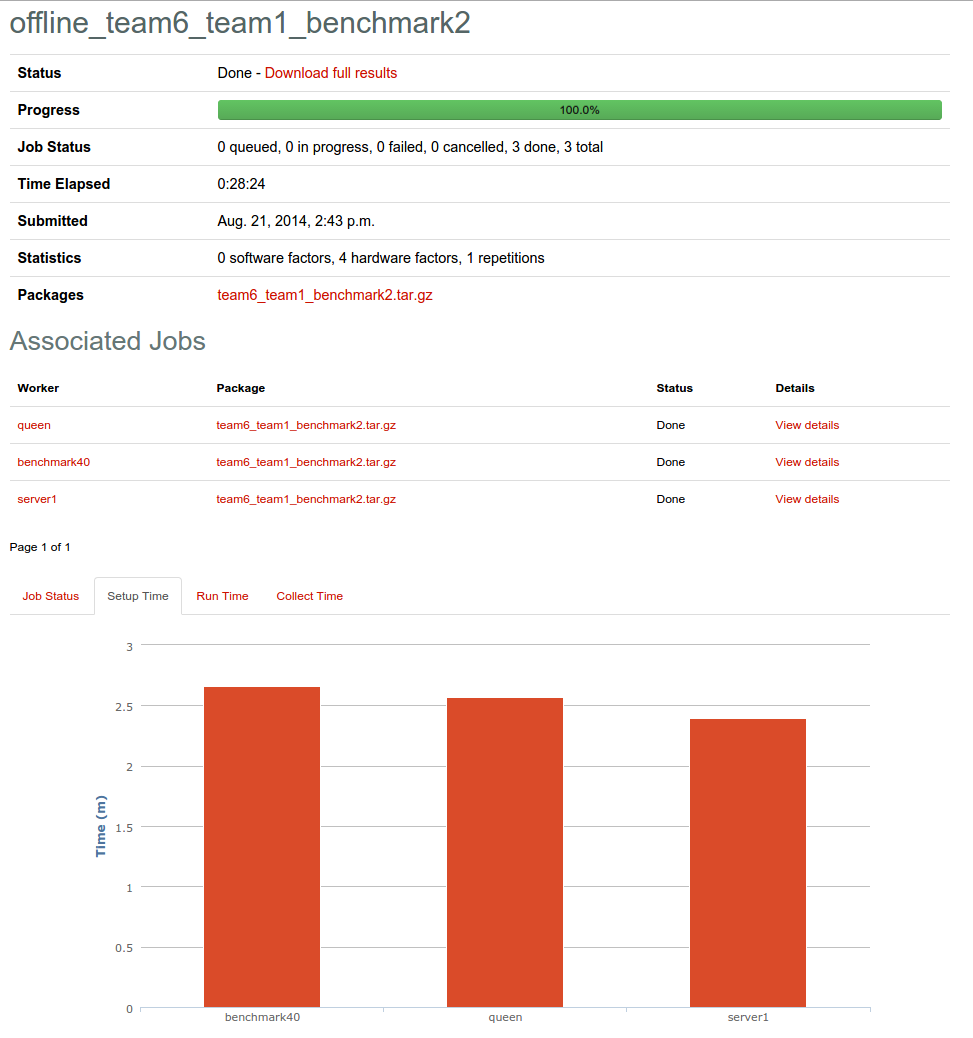
After the jobs complete, the experiment page will be populated with data and statistics about the process. It will give the times for each of the three phases (setup, run, collect) for each replicated run. It will also allow you to see breakdowns of each replicated job. You can download the original uploaded package and also download a tar with the collected results from all jobs.
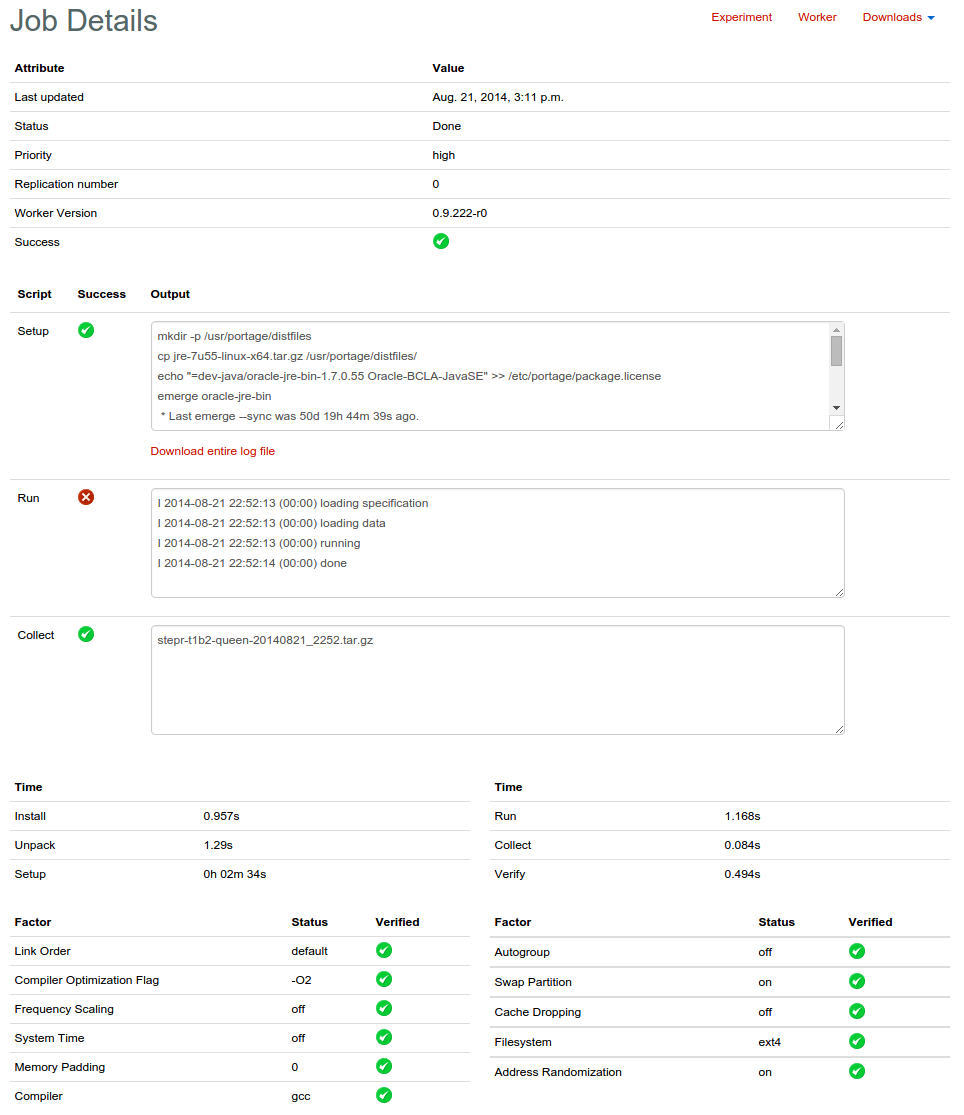
The page for an individual run will show you the output of each of the three phases and whether or not that phase succeeded. It gives you a breakdown of what was installed on the worker, what the kernel is, and any other factors that were 1. requested and 2. available on the machine.
From there, you can share your experiment to anyone with an account on DataMill. There is no way to publicly share the experiment with anybody else. You can fork/clone the experiment by downloading the package and reuploading it yourself as your own experiment. The relationship and thus provenance, however, is lost.
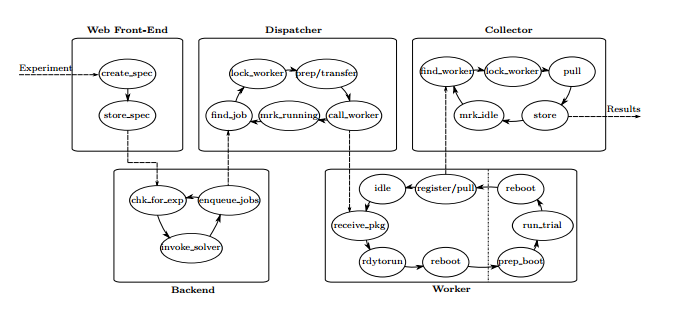
Infrastructure
There are a set of worker nodes that coordinate with a master node. These worker nodes are provided by the community from places such as Purdue University and the University of Pennsylvania (according to section 4.2 of their paper.) These machines run Gentoo and a copy of the DataMill worker code.
The worker nodes are manually established to have two partitions: a message partition that receives commands and a boot partition that will run the actual experiment. This is so different flags and parameters can be established for the experiment and can control for any impact of the message system. The system, when it receives a command to run an experiment and a payload, will reboot the system into the boot system which will run the scripts and then reboot once more back to the message partition.
Currently there are more than 25 machines available. They represent various ARM and x86-64 (i586/Xeon/etc) chipsets and various GPU architectures. DataMill will add workers as people from other institutions allow.
Capabilities
Access
All views of experiments and data require an active DataMill account to view. Therefore, you cannot easily share the work or any description of that work through the DataMill service to any collaborators nor reviewers. You cannot easily link to the experiment from a paper for the same reason. You can download the generated files, however, and share those.
Provenance
The experiments cannot pull information from other experiments nor the world at large. The exception is any network activity that may happen in a setup script. Some workers may not allow some network activity due to differences in environment or firewall configurations. Since there is no way to fork or clone, related experiments may not be recorded unless tagged carefully.
Within the DataMill network, some relationships are known. An experiment is tagged with its author and the time and date it was executed. It knows what type of hardware and which machine it was executed upon. It keeps track of the libraries resident on the machine along with the flags and logs of its setup process and its runtime.
Therefore, it is strong hardware provenance and weaker relational provenance and data origin provenance.
Governance
Strengths
Breakdown
- Easily dispatch on a diverse set of hardware
- Simple to wrap experimentation
- Keeps track of hardware and system environment parameters
Weaknesses
Breakdown
- Difficult to self-host and therefore centralized
- Workers are constrained
- No public access; open moderated accounts
Unique Features
To be discussed.
Best-Practice Influences
To be discussed.
Digital Library Incorporation Issues
To be discussed.